You can do more for neural networks in R with {kindling}

This post has been written in collaboration with Joshua Marie.
Why this post matters
Neural networks in R are no longer niche. Today, we can choose among:
{nnet}for classic, small-scale neural nets,{neuralnet}another classic neural nets package besides{nnet},{keras}/{keras3}for the Keras API (typically with Python backends such as TensorFlow/JAX/Torch),{torch}for native-R deep learning with explicit model and training control.
So why discuss another package?
In our experience, many day-to-day projects sit in the middle: we want more flexibility than a classical model, but less boilerplate than writing a full {torch} training loop. {kindling} is designed for that middle ground.
In this article, we focus on what {kindling} does well, where it helps, and where it is still limited.
What problem {kindling} solves (and what it does not)
{kindling} is a higher-level interface built on top of {torch}. In practice, it reduces repetitive code for:
- defining common network architectures,
- training feedforward and recurrent models,
- integrating with
{tidymodels}({parsnip},{recipes},{workflows},{tune}).
It does not replace low-level {torch} for highly customized research code, and it does not make hardware setup disappear. You still need a working LibTorch installation and an environment that can use CPU/GPU properly.
At the time of writing (CRAN version 0.2.0), core built-in architectures include FFNN/MLP and recurrent variants (RNN/LSTM/GRU).
Setup
Install this package in two different ways:
- Solely install
{kindling}(as it installs the package dependencies internally)
install.packages("kindling")- You can install the package dependencies separately:
install.packages(c(
"kindling",
# dependencies
"torch", "dplyr", "rsample", "recipes", "yardstick",
"workflows", "parsnip", "tibble", "ggplot2", "mlbench", "vip"
))Then load the packages in two ways:
- Using
library()traditionally:
library(kindling)
library(torch)
library(dplyr)
library(rsample)
library(recipes)
library(yardstick)
library(workflows)
library(parsnip)
library(tibble)
library(ggplot2)
library(mlbench)
library(vip)In this article, we use library() for clarity and copy-paste reproducibility.
- If you prefer a module-based and more explicit import style, use
box::use()(read Box: Placing module system into R for more details).
Here’s the equivalent code as above:
box::use(
kindling[...],
dplyr[...],
rsample[...],
recipes[...],
yardstick[...],
workflows[...],
parsnip[...],
tibble[...],
ggplot2[...],
mlbench[...],
vip[...]
){kindling} uses {torch} as backend, so LibTorch must be installed once:
if (!torch::torch_is_installed()) {
torch::install_torch()
}Three levels of interaction
{kindling} is useful because you can work at different abstraction levels. Here are three ways to interact with the package, ordered from lowest to highest level of abstraction:
1) Generate model code (*_generator())
If you want to inspect architecture code before training:
ffnn_generator(
nn_name = "MyNet",
hd_neurons = c(64, 32, 16),
no_x = 10,
no_y = 1,
activations = act_funs(
relu,
"softplus(beta = 0.5)",
selu
)
)## torch::nn_module("MyNet", initialize = function ()
## {
## self$fc1 = torch::nn_linear(10, 64, bias = TRUE)
## self$fc2 = torch::nn_linear(64, 32, bias = TRUE)
## self$fc3 = torch::nn_linear(32, 16, bias = TRUE)
## self$out = torch::nn_linear(16, 1, bias = TRUE)
## }, forward = function (x)
## {
## x = self$fc1(x)
## x = torch::nnf_relu(x)
## x = self$fc2(x)
## x = torch::nnf_softplus(x, beta = 0.5)
## x = self$fc3(x)
## x = torch::nnf_selu(x)
## x = self$out(x)
## x
## })Technical note: To specify a parametric activation functions like Softplus under act_funs() function, set softplus = args(beta = 0.5) or softplus[beta = 0.5] (available in v0.3.x and later), not just in a stringly typed expression, e.g. "softplus(beta = 0.5)".
This returns an unevaluated torch::nn_module expression you can inspect or modify.
2) Direct Training
The functions available (for now) are ffnn() and rnn(). For many applied tasks, this is the fastest way to fit a network from a formula.
mini_direct <- ffnn(
mpg ~ .,
data = mtcars,
hidden_neurons = 8,
activations = act_funs(relu),
epochs = 20,
verbose = FALSE
)
predict(mini_direct, newdata = head(mtcars, 3))## [1] 11.21900 11.21385 10.568413) ML Framework Integration: {tidymodels} with mlp_kindling() / rnn_kindling()
{kindling} functions to directly train the said models also (currently) integrates with {tidymodels}. This level is ideal when we want recipes, workflows, resampling, and tuning.
mini_spec <- mlp_kindling(
mode = "regression",
hidden_neurons = 8,
activations = act_funs(relu), # or just "relu"
epochs = 20,
verbose = FALSE
)
mini_wf <- workflow() |>
add_formula(mpg ~ .) |>
add_model(mini_spec)
mini_fit <- fit(mini_wf, data = mtcars)
predict(mini_fit, new_data = head(mtcars, 3))## # A tibble: 3 × 1
## .pred
## <dbl>
## 1 -3.49
## 2 -3.50
## 3 -3.50Now that we’ve covered the three levels of interaction, let’s see {kindling} in action with two “realistic” examples. We’ll demonstrate how to structure reproducible workflows, handle feature preprocessing, and evaluate results properly. Along the way, you’ll see which abstraction level works best for different scenarios.
Example 1: Iris classification with reproducible good practice
This first example uses direct training with ffnn(), but still follows practical steps:
- train/test split,
- feature scaling,
- validation split during training,
- regularization,
- out-of-sample evaluation.
Data Preprocessing
Train/test split is performed to prevent evaluating only on training data.
data(iris)
iris_split <- initial_split(iris, prop = 0.8, strata = Species)
iris_train <- training(iris_split)
iris_test <- testing(iris_split)
iris_recipe <- recipe(Species ~ ., data = iris_train) |>
step_normalize(all_predictors())
iris_prep <- prep(iris_recipe, training = iris_train)
iris_train_processed <- bake(iris_prep, new_data = NULL)
iris_test_processed <- bake(iris_prep, new_data = iris_test)Example 2: A more realistic tabular benchmark (Sonar)
Iris is useful for teaching, but sometimes considered (too) easy. The Sonar dataset (60 numeric predictors, binary outcome) is a better stress test for tabular classification.
data("Sonar", package = "mlbench")
sonar <- Sonar |>
mutate(Class = factor(Class))
sonar_split <- initial_split(sonar, prop = 0.8, strata = Class)
sonar_train <- training(sonar_split)
sonar_test <- testing(sonar_split)Now fit through {tidymodels} so preprocessing and modeling stay in one pipeline:
sonar_rec <- recipe(Class ~ ., data = sonar_train) |>
step_normalize(all_predictors())
sonar_spec <- mlp_kindling(
mode = "classification",
hidden_neurons = c(64, 32),
activations = act_funs(relu, relu),
optimizer = "adam",
learn_rate = 0.001,
penalty = 1e-4,
mixture = 0,
batch_size = 16,
epochs = 250,
validation_split = 0.2,
verbose = FALSE
)
sonar_wf <- workflow() |>
add_recipe(sonar_rec) |>
add_model(sonar_spec)
set.seed(20260218)
sonar_fit <- fit(sonar_wf, data = sonar_train)sonar_pred <- augment(sonar_fit, new_data = sonar_test)
metrics(sonar_pred, truth = Class, estimate = .pred_class)## # A tibble: 2 × 3
## .metric .estimator .estimate
## <chr> <chr> <dbl>
## 1 accuracy binary 0.837
## 2 kap binary 0.672In practice, this setup is often a strong baseline for tabular binary classification before trying larger architectures.
About early stopping and callbacks
If you come from Keras/TensorFlow, you may be used to callback objects (early stopping, learning-rate schedules, etc.).
With {kindling} 0.2.0, practical overfitting control is usually handled through:
validation_split,- regularization (
penalty,mixture), - tuning model size and training length (
epochs), - proper resampling with
{tidymodels}.
In other words, we can approximate early-stopping behavior by selecting epochs via validation/resampling, even without a callback-heavy workflow.
Supported architectures (current scope)
| Architecture | Main function(s) | Typical use |
|---|---|---|
| Feedforward (MLP/FFNN) | ffnn(), mlp_kindling() | Tabular regression/classification |
| RNN | rnn_kindling(rnn_type = "rnn") | Sequential patterns |
| LSTM | rnn_kindling(rnn_type = "lstm") | Longer-range sequence dependencies |
| GRU | rnn_kindling(rnn_type = "gru") | Sequence modeling with fewer parameters |
Variable importance (FFNN)
Interpretability for neural networks is imperfect, but {kindling} integrates established approaches for FFNN models.
# For FFNN fits:
garson(iris_mlp, bar_plot = FALSE)## x_names y_names rel_imp
## 1 Petal.Length y 33.39467
## 2 Sepal.Width y 24.44282
## 3 Petal.Width y 21.92705
## 4 Sepal.Length y 20.23546olden(iris_mlp, bar_plot = FALSE)## x_names y_names rel_imp
## 1 Petal.Length y -2.4176215
## 2 Petal.Width y -1.8339908
## 3 Sepal.Length y -0.6675719
## 4 Sepal.Width y 0.5844613# Via vip (Olden/Garson methods supported by kindling S3 methods)
vi(iris_mlp, type = "olden") |>
vip()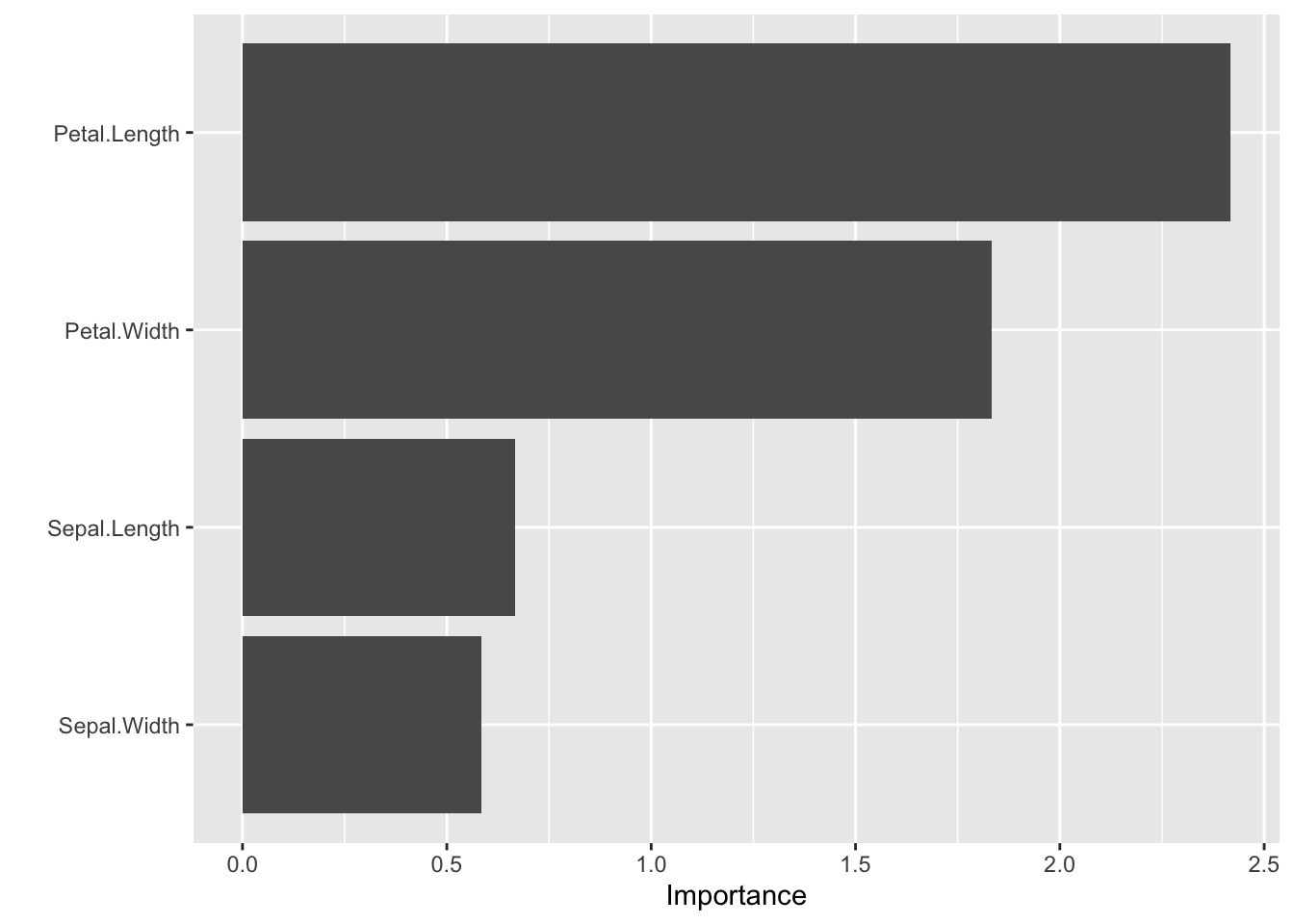
We recommend using these as directional diagnostics, not as causal evidence.
When {kindling} is a good fit
{kindling} is a practical choice when our projects:
- work mostly in R and want to stay inside
{tidymodels}, - need neural nets for tabular or moderate sequence tasks,
- want less boilerplate than raw
{torch}but still meaningful control.
It may be less ideal when our projects need:
- custom research architectures and training loops,
- mature callback ecosystems similar to high-level Keras workflows,
- highly optimized distributed production pipelines.
Limitations to keep in mind
- Hardware setup still matters: GPU usage depends on a correct
{torch}/LibTorch installation and supported hardware. - Ecosystem maturity: this is a young package; interfaces can evolve.
- Debugging depth: for deeply custom debugging, low-level
{torch}remains the reference path. - Model class assumptions: recurrent models are for sequence-structured data; using them on plain tabular data is usually not appropriate.
Takeaways
{kindling} is not about replacing {torch} or {keras3}. It is about reducing friction for common deep-learning workflows in R.
For many applied projects, a robust pattern is:
- preprocess with
{recipes}, - start with a modest MLP architecture,
- use validation split + regularization,
- evaluate on held-out test data,
- tune only after you have a strong baseline.
If this matches your workflow, {kindling} is worth trying.
As always, if you have any question related to the topic covered in this post, please add it as a comment so other readers can benefit from the discussion.
Liked this post?
- Get updates every time a new article is published (no spam and unsubscribe anytime)
- Support the blog
- Share on:



